IIT and EMBL scientists reveal detailed insights into self-splicing introns
Computer simulations and X-ray protein crystallography worked together to reveal insight into the mechanism of gene splicing. Researchers at IIT in Genova, led by Marco De Vivo (De Vivo lab), and EMBL in Grenoble, led by Marco Marcia (Marcia group), have obtained some of the most detailed ever snapshots of the splicing process in systems known as group II self-splicing introns, that is specific part of RNA. Their findings are published in Nature Communications (https://www.nature.com/articles/s41467-020-16741-4) and may have implications for biotechnology and drug development.
Marco De Vivo, Head at IIT’s Molecular Modeling and Drug Discovery Lab told us that “these findings come from a few years of intense work, integrating advanced molecular simulations with experimental work to explain such fundamental biological process regulated by an RNA enzyme”. This kind of study are fundamental for designing new drugs and gene-specific therapeutics, because they permit to reveal new mechanism to be used or targeted to resolve specific genetic or infectious diseases. “Indeed, this has been a great teamwork of our labs, at EMBL and IIT – Marco and I look forward to continuing our collaborative efforts on this exciting line of research”, remarks Marco Marcia in Grenoble.
The research study involved two young researchers, Jacopo Manigrasso, PhD student in the De Vivo lab, and Isabel Chillón, Postdoctoral Fellow in the Marcia group, who are the first authors of the study. Results are described on EMBL News website (https://www.embl.org/news/science/genetic-cut-and-paste-at-atomic-resolution/)
Computer simulations were used to reproduce at atomic scale the splicing process carried out by a molecular machine called the spliceosome, which is a complex assembly of RNA molecules supported by proteins.“These self-splicing introns, called group II introns, are the evolutionary ancestors of the RNA units of the spliceosome, and the two machineries use the very same chemical reaction to perform splicing,” explains Jacopo Manigrasso. “By understanding the self-splicing mechanism of group II introns, we can better understand how splicing works in humans as well.”
Splicing is a crucial step in using the genetic information stored in DNA to make proteins, which carry out the majority of biological functions. To make genetic information usable, DNA has to be converted, or transcribed, into messenger RNA (mRNA) – a molecule that acts as a transient carrier of information. In animals, most genes and their mRNA transcripts contain long sequences of genetic information that are not used directly to make part of a protein. These segments, which are called introns, have to be cut from their mRNA strand before it can be used to produce a functional protein. By cutting different combinations of introns from an mRNA, cells can also produce different protein variants from a single gene. This allows organisms to increase the number of proteins they can make, without having to carry a separate gene for each protein, keeping the genome compact.
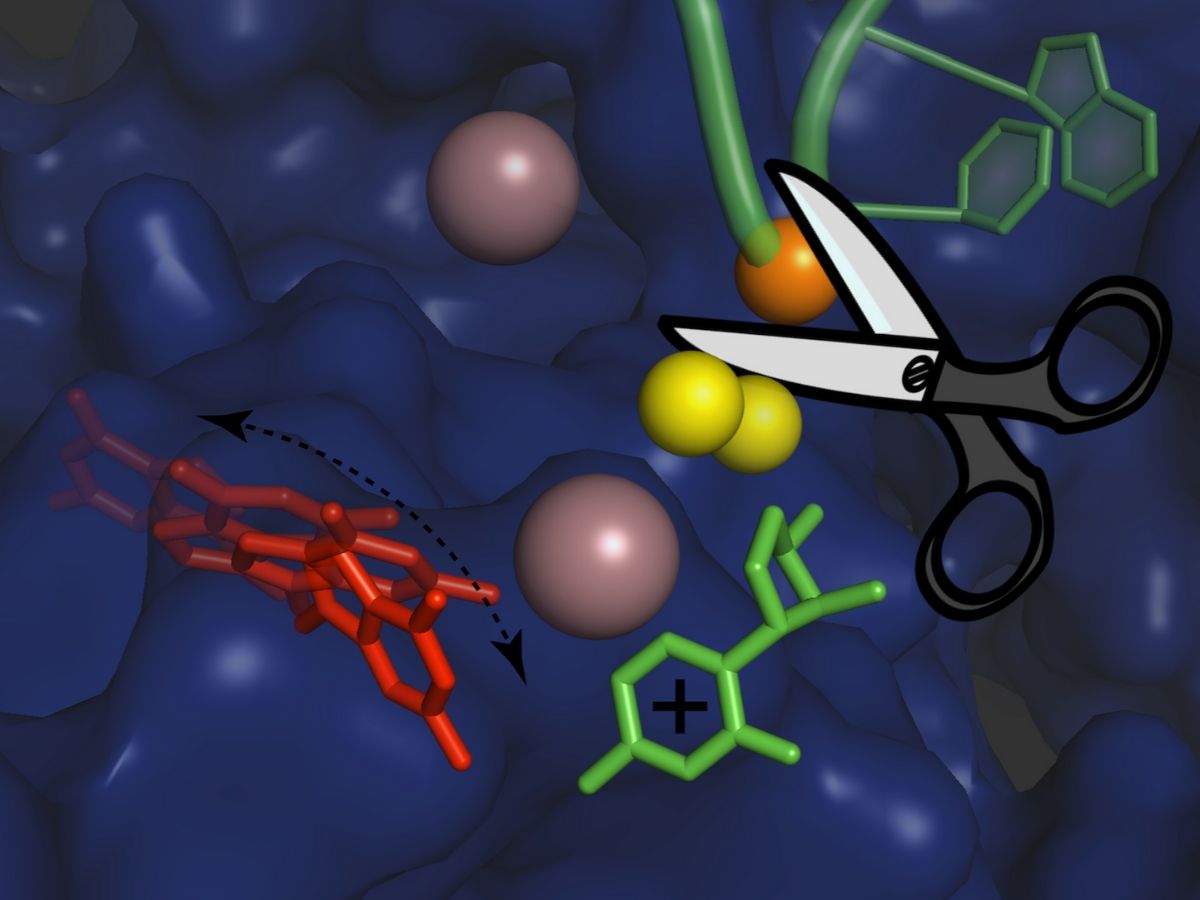
Researchers used X-ray protein crystallography and computer simulations to figure out the atomic details of the group II intron splicing reaction. “Each splicing event consists of two major steps: a cut at one end of an intron, followed by a cut at the other end that releases the intron and simultaneously reconnects the two flanking exons of the mRNA,” explains Isabel Chillón. “In between these steps, the intron needs to be slightly rearranged – but the precise mechanism of this rearrangement has not been known until now.”
The scientists have now found that the first step of the splicing reaction unleashes a positive charge within the intron. This leads to the temporary detachment of a potassium ion, which assists splicing and stabilises the intron structure, and causes a flip of the intron in the proximity of the cutting site. This transient rearrangement is crucial to establish a suitable configuration for the intron to perform the second step of the splicing reaction. When the scientists prevented the positive charge transfer in their experiments, they found that the sequence of rearrangements does not take place, and the second ‘cut and paste’ step of the splicing reaction happens at an impractically slow rate.
Future applications will be in the development of small molecules, called splicing inhibitors, which selectively block group II intron splicing without affecting the spliceosome. Since self-splicing introns are essential for the survival of certain pathogens, such inhibitors could in the future be used to treat infectious diseases, expanding our toolkit of antibiotics.